Stationarity tests
1 Dickey–Fuller test
The Dickey–Fuller test tests the null hypothesis that a unit root is present in an autoregressive (AR) model. The alternative hypothesis is different depending on which version of the test is used, but is usually stationarity or trend-stationarity. Let’s consider an AR(1) model, i.e.
The, the Dickey–Fuller hypothesis are:
2 Augmented Dickey–Fuller test
The augmented Dickey–Fuller is a more general version of the Dickey–Fuller test for a general AR(p) model, i.e.
Then, the augmented Dickey–Fuller hypothesis are:
3 Kolmogorov-Smirnov test
The Kolmogorov–Smirnov two-sample test (KS test) can be used to test whether two samples came from the same distribution. Let’s define the empirical distribution function
To apply the test in a time series settings, it is possible to random split the original series in two sub samples and apply the test above.
3.1 Example: check stationary
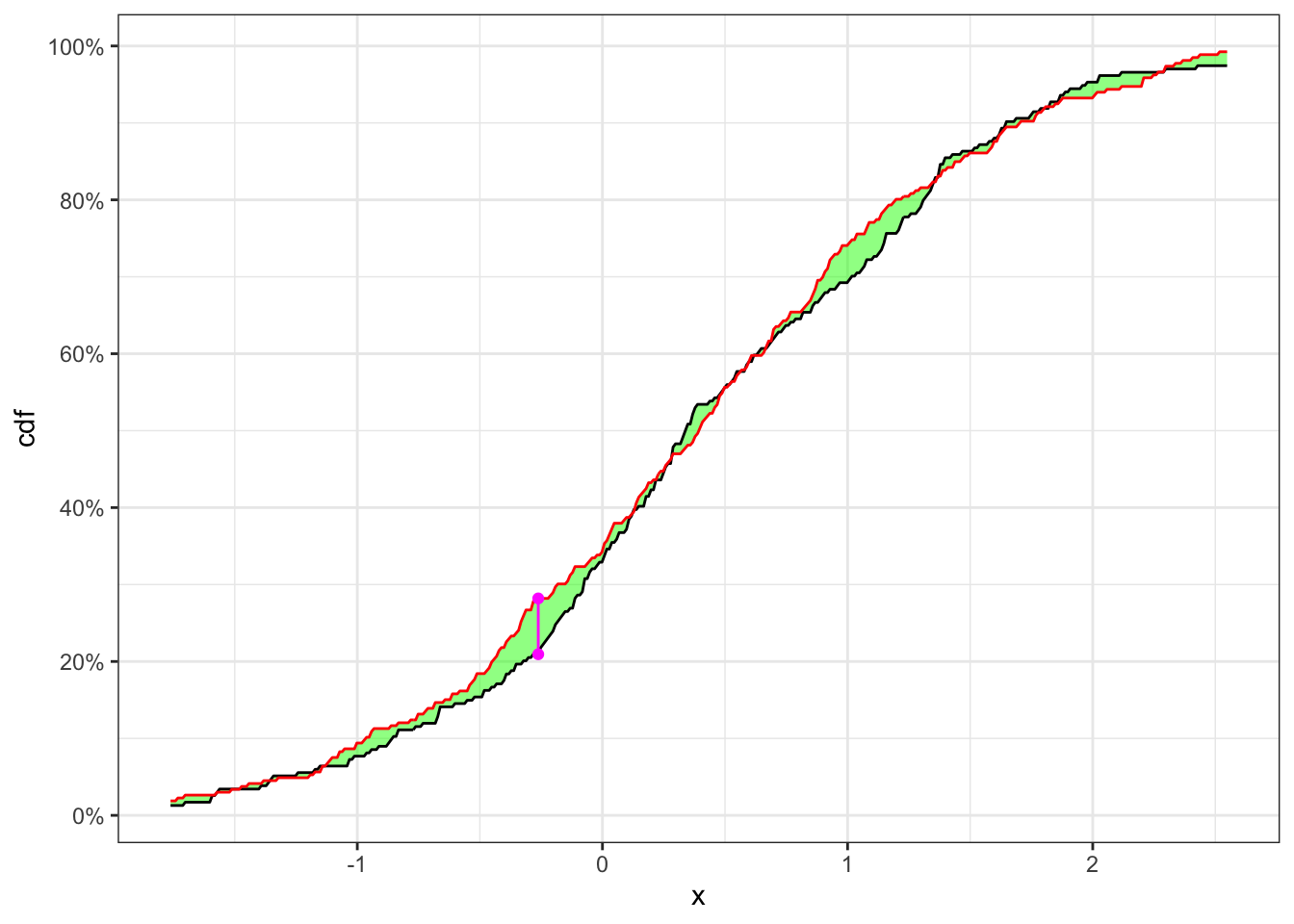
Let’s simulate 500 observations of
KS-test on a stationary time series
# ============== Setups ==============
set.seed(5) # random seed
ci <- 0.05 # confidence level (alpha)
n <- 500 # number of simulations
# ====================================
# Simulated stationary sample
x <- rnorm(n, 0.4, 1)
# Random split of the time series
idx_split <- sample(n, 1)
x1 <- x[1:idx_split]
x2 <- x[(idx_split+1):n]
# Number of elements for each sub sample
n1 <- length(x1)
n2 <- length(x2)
# Grid of values for KS-statistic
grid <- seq(quantile(x, 0.015), quantile(x, 0.985), 0.01)
# Empiric cdfs
cdf_1 <- ecdf(x1)
cdf_2 <- ecdf(x2)
# KS-statistic
ks_stat <- max(abs(cdf_1(grid) - cdf_2(grid)))
# Rejection level with probability alpha
rejection_lev <- sqrt(-0.5*log(ci/2))*sqrt((n1+n2)/(n1*n2))
# ========================== Plot ==========================
y_breaks <- seq(0, 1, 0.2)
y_labels <- paste0(format(y_breaks*100, digits = 2), "%")
grid_max <- grid[which.max(abs(cdf_1(grid) - cdf_2(grid)))]
ggplot()+
geom_ribbon(aes(grid, ymax = cdf_1(grid), ymin = cdf_2(grid)),
alpha = 0.5, fill = "green") +
geom_line(aes(grid, cdf_1(grid)))+
geom_line(aes(grid, cdf_2(grid)), color = "red")+
geom_segment(aes(x = grid_max, xend = grid_max,
y = cdf_1(grid_max), yend = cdf_2(grid_max)),
linetype = "solid", color = "magenta")+
geom_point(aes(grid_max, cdf_1(grid_max)), color = "magenta")+
geom_point(aes(grid_max, cdf_2(grid_max)), color = "magenta")+
scale_y_continuous(breaks = y_breaks, labels = y_labels)+
labs(x = "x", y = "cdf")+
theme_bw()In Table 1 the null hypothesis, i.e. the two samples come from the same distribution, is not reject with the confidence level
3.2 Example: check non-stationary
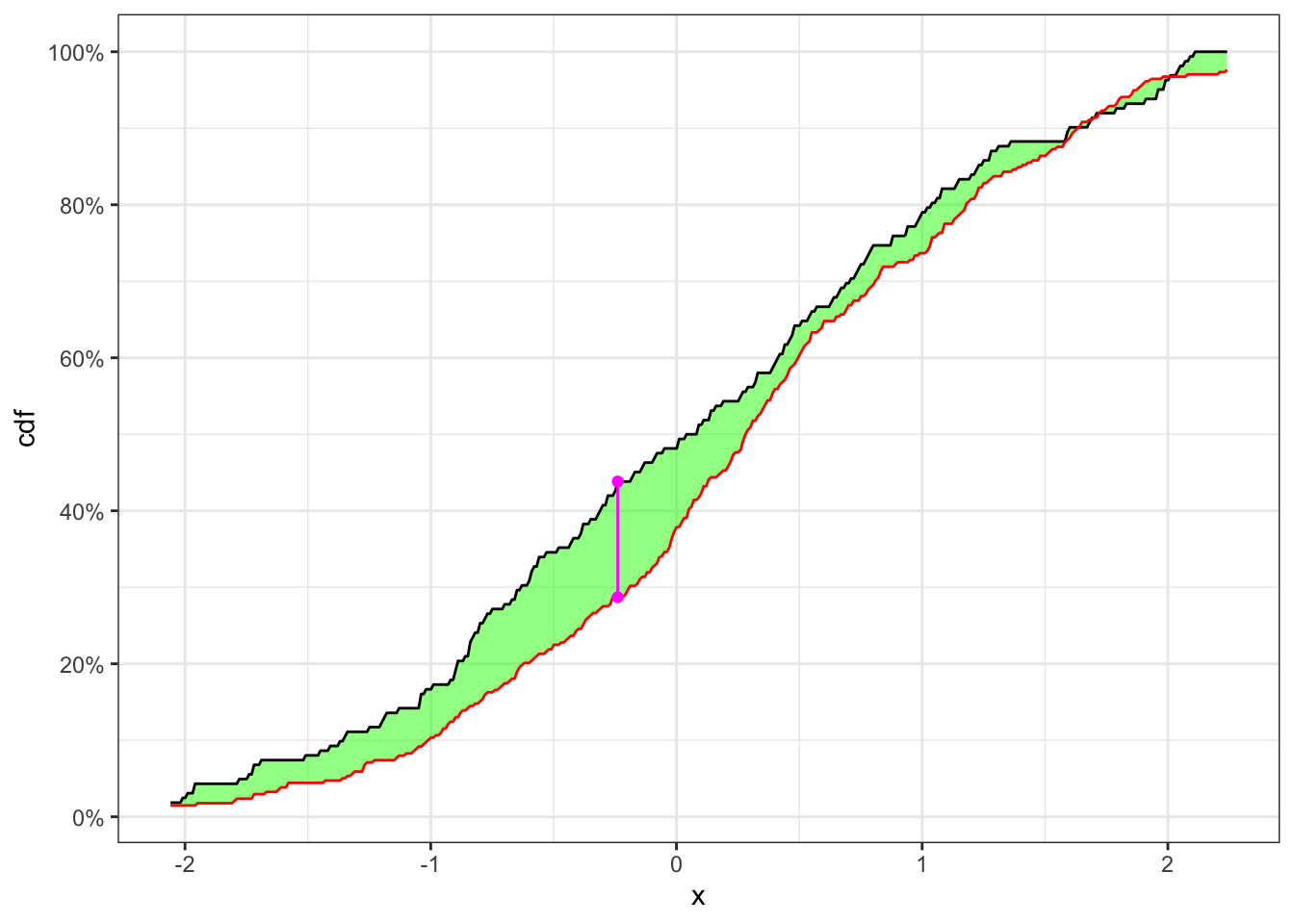
Let’s now simulate 250 observations as
KS-test on a non-stationary time series
# ============== Setups ==============
set.seed(2) # random seed
ci <- 0.05 # confidence level (alpha)
n <- 500 # number of simulations
# ====================================
# Simulated non-stationary sample
x1 <- rnorm(n/2, 0, 1)
x2 <- rnorm(n/2, 0.3, 1)
x <- c(x1, x2)
# Random split of the time series
idx_split <- sample(n, 1)
x1 <- x[1:idx_split]
x2 <- x[(idx_split+1):n]
# Number of elements for each sub sample
n1 <- length(x1)
n2 <- length(x2)
# Grid of values for KS-statistic
grid <- seq(quantile(x, 0.015), quantile(x, 0.985), 0.01)
# Empiric cdfs
cdf_1 <- ecdf(x1)
cdf_2 <- ecdf(x2)
# KS-statistic
ks_stat <- max(abs(cdf_1(grid) - cdf_2(grid)))
# Rejection level
rejection_lev <- sqrt(-0.5*log(ci/2))*sqrt((n1+n2)/(n1*n2))
# ========================== Plot ==========================
y_breaks <- seq(0, 1, 0.2)
y_labels <- paste0(format(y_breaks*100, digits = 2), "%")
grid_max <- grid[which.max(abs(cdf_1(grid) - cdf_2(grid)))]
ggplot()+
geom_ribbon(aes(grid, ymax = cdf_1(grid), ymin = cdf_2(grid)),
alpha = 0.5, fill = "green") +
geom_line(aes(grid, cdf_1(grid)))+
geom_line(aes(grid, cdf_2(grid)), color = "red")+
geom_segment(aes(x = grid_max, xend = grid_max,
y = cdf_1(grid_max), yend = cdf_2(grid_max)),
linetype = "solid", color = "magenta")+
geom_point(aes(grid_max, cdf_1(grid_max)), color = "magenta")+
geom_point(aes(grid_max, cdf_2(grid_max)), color = "magenta")+
scale_y_continuous(breaks = y_breaks, labels = y_labels)+
labs(x = "x", y = "cdf")+
theme_bw()In Table 2 the null hypothesis, i.e. the two samples come from the same distribution, is reject with a confidence level
Citation
@online{sartini2024,
author = {Sartini, Beniamino},
title = {Stationarity Tests},
date = {2024-05-01},
url = {https://greenfin.it/statistics/tests/stationarity-tests.html},
langid = {en}
}