ARCH-GARCH process
1 ARCH(p) process
The autoregressive conditional heteroskedasticity (ARCH) models were introduced in 1982 by Robert Engle to model varying (conditional) variance of time series. It is often found in economics that the larger values of time series also lead to larger instability. Let’s define a time series such as
The unconditional and conditional mean of a process
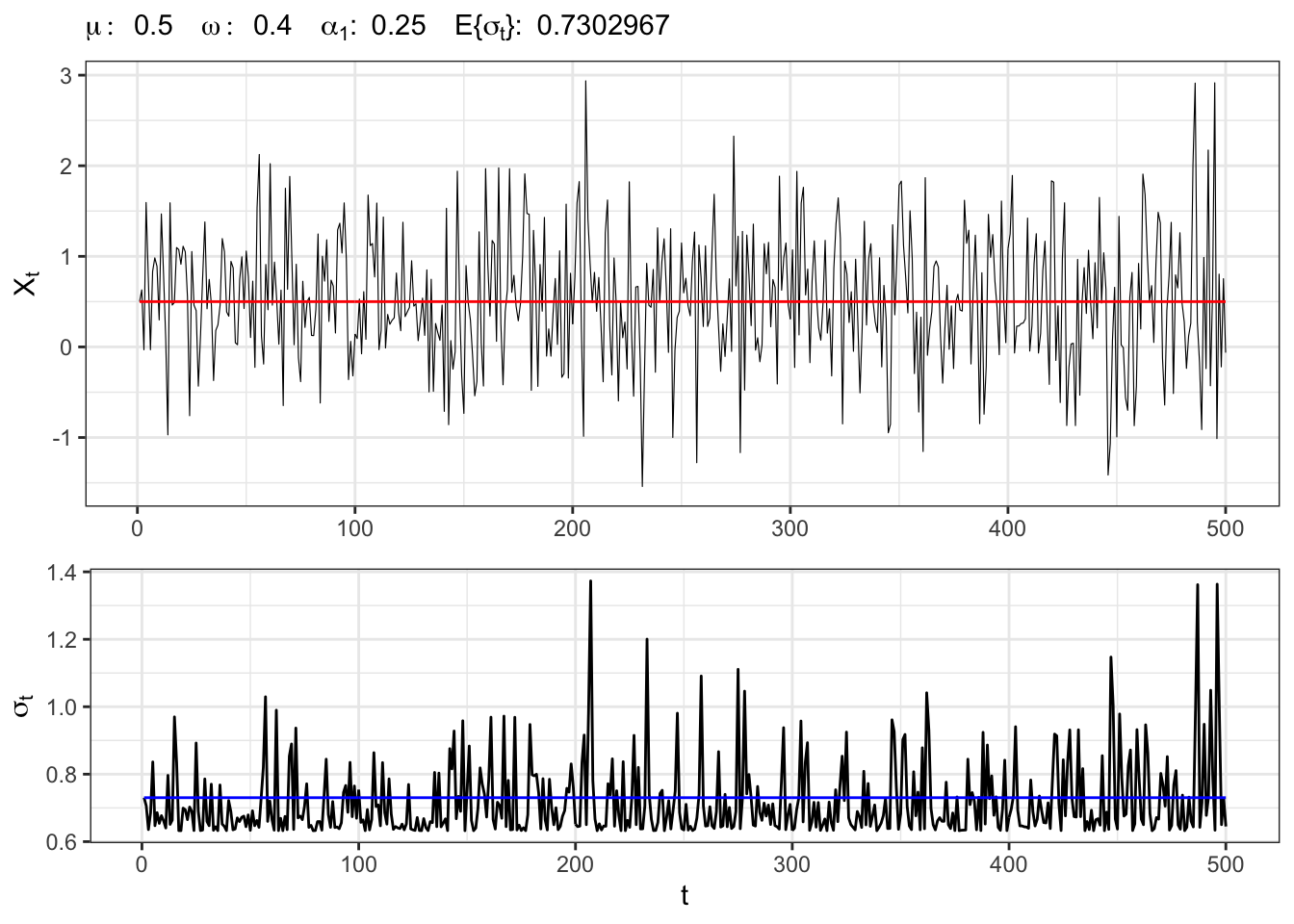
1.1 Example: ARCH(1) process
Let’s simulate an ARCH(1) process with normal residuals, namely
ARCH(1) simulation
# Random seed
set.seed(1)
# ==================== Setups =====================
# Nsumber of simulations
t_bar <- 500
# Parameters
par <- c(mu=0.5, omega=0.4, alpha1=0.25)
# Long-term std deviation of residuals
sigma_eps <- sqrt(par[2]/(1-par[3]))
# ================== Simulation ===================
# Initial point
Xt <- rep(par[1], 1)
# Store stochastic variance
sigma <- rep(sigma_eps, 1)
# Simulated residuals
eps <- rnorm(t_bar, 0, 1)
for(t in 2:t_bar){
# ARCH(1) variance
sigma[t] <- par[3]*eps[t-1]^2
# ARCH(1) std. deviation
sigma[t] <- sqrt(par[2] + sigma[t])
# Simulated residuals
eps[t] <- sigma[t]*eps[t]
# Simulated time series
Xt[t] <- par[1] + eps[t]
}
# ===================== Plot ======================
# ARCH(1) simulation
plot_arch <- ggplot()+
geom_line(aes(1:t_bar, Xt), size = 0.2)+
geom_line(aes(1:t_bar, par[1]), color = "red")+
theme_bw()+
labs(x = NULL, y = TeX("$X_t$"),
subtitle = TeX(paste0("$\\mu:\\;", par[1],
"\\;\\; \\omega:\\;", par[2],
"\\;\\; \\alpha_{1}:\\;", par[3],
"\\;\\; E\\{\\sigma_t\\}:\\;", sigma_eps,
"$")))
# ARCH(1) std. deviation
plot_sigma <- ggplot()+
geom_line(aes(1:t_bar, sigma), alpha = 1)+
geom_line(aes(1:t_bar, sigma_eps), alpha = 1, color = "blue")+
labs(x = "t", y = TeX("$\\sigma_{t}$"))+
theme_bw()
gridExtra::grid.arrange(plot_arch, plot_sigma, heights = c(0.6, 0.4))1.2 Example: ARCH(3) process
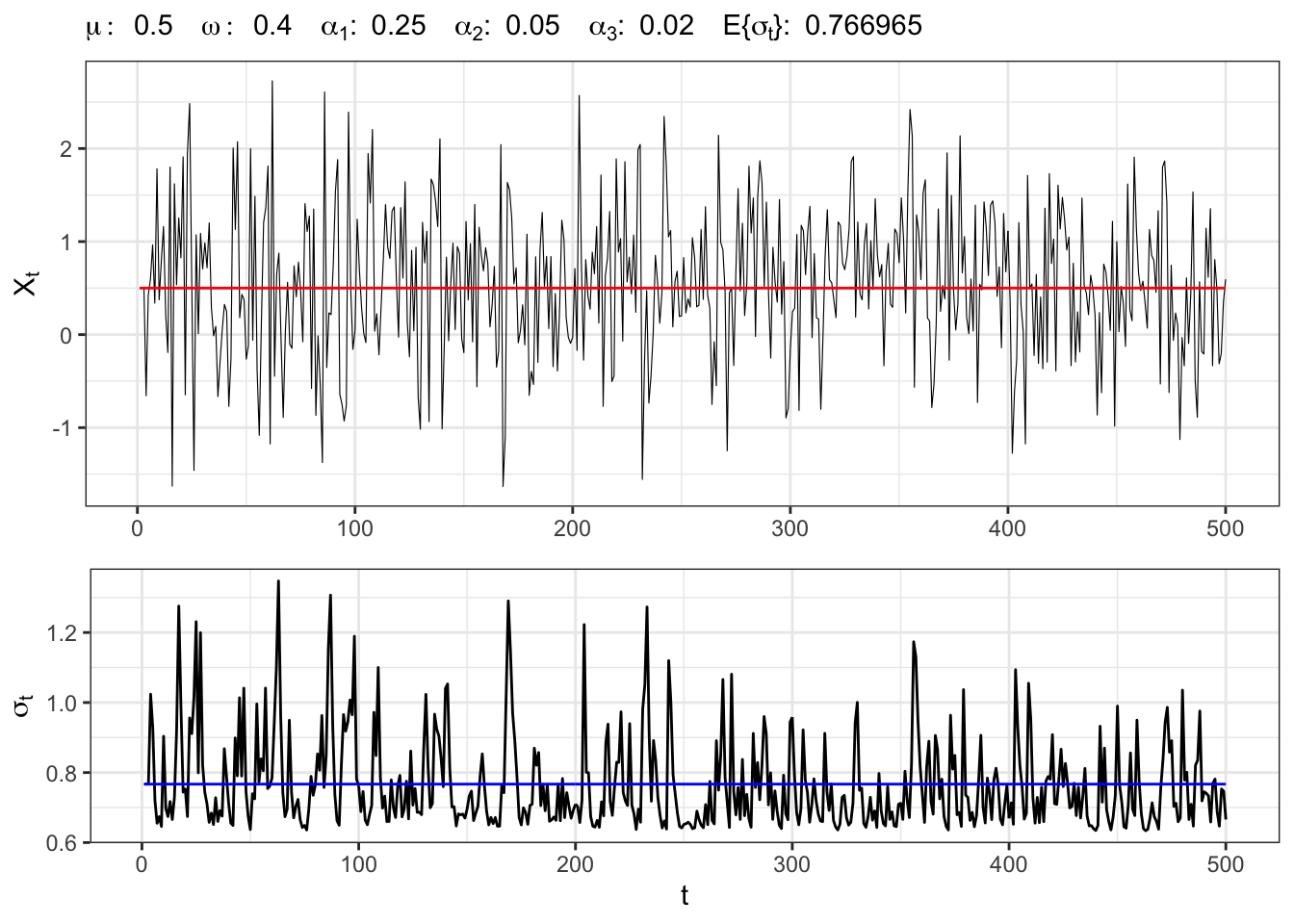
Let’s simulate an ARCH(3) process with normal residuals, namely
ARCH(3) simulation
# Random seed
set.seed(2)
# ==================== Setups =====================
# Number of simulations
t_bar <- 500
# Parameters
par <- c(mu=0.5, omega=0.4,
alpha1=0.25, alpha2=0.05,alpha3=0.02)
# Long-term std deviation of residuals
sigma_eps <- sqrt(par[2]/(1-sum(par[3:5])))
# ================== Simulation ===================
# Initial point
Xt <- rep(par[1], 3)
# Store stochastic variance
sigma <- rep(sigma_eps, 3)
# Simulated residuals
eps <- rnorm(t_bar, 0, 1)
for(t in 4:t_bar){
# ARCH(3) variance
sigma[t] <- par[3]*eps[t-1]^2 + par[4]*eps[t-2]^2 + par[5]*eps[t-3]^2
# ARCH(3) std. deviation
sigma[t] <- sqrt(par[2] + sigma[t])
# Simulated residuals
eps[t] <- sigma[t]*eps[t]
# Simulated time series
Xt[t] <- par[1] + eps[t]
}
# ===================== Plot ======================
# ARCH(3) simulation
plot_arch <- ggplot()+
geom_line(aes(1:t_bar, Xt), size = 0.2)+
geom_line(aes(1:t_bar, par[1]), color = "red")+
theme_bw()+
labs(x = NULL, y = TeX("$X_t$"),
subtitle = TeX(paste0("$\\mu:\\;", par[1],
"\\;\\; \\omega:\\;", par[2],
"\\;\\; \\alpha_{1}:\\;", par[3],
"\\;\\; \\alpha_{2}:\\;", par[4],
"\\;\\; \\alpha_{3}:\\;", par[5],
"\\;\\; E\\{\\sigma_t\\}:\\;", sigma_eps,
"$")))
# ARCH(3) std. deviation
plot_sigma <- ggplot()+
geom_line(aes(1:t_bar, sigma), alpha = 1)+
geom_line(aes(1:t_bar, sigma_eps), alpha = 1, color = "blue")+
labs(x = "t", y = TeX("$\\sigma_{t}$"))+
theme_bw()
gridExtra::grid.arrange(plot_arch, plot_sigma, heights = c(0.6, 0.4))2 GARCH(p,q) process
As done with the ARCH(p), with generalized autoregressive conditional heteroskedasticity (GARCH) we model the dependency of the conditional second moment. It represents a more parsimonious way to express the conditional variance. Let’s consider a process
The unconditional and conditional mean of a process
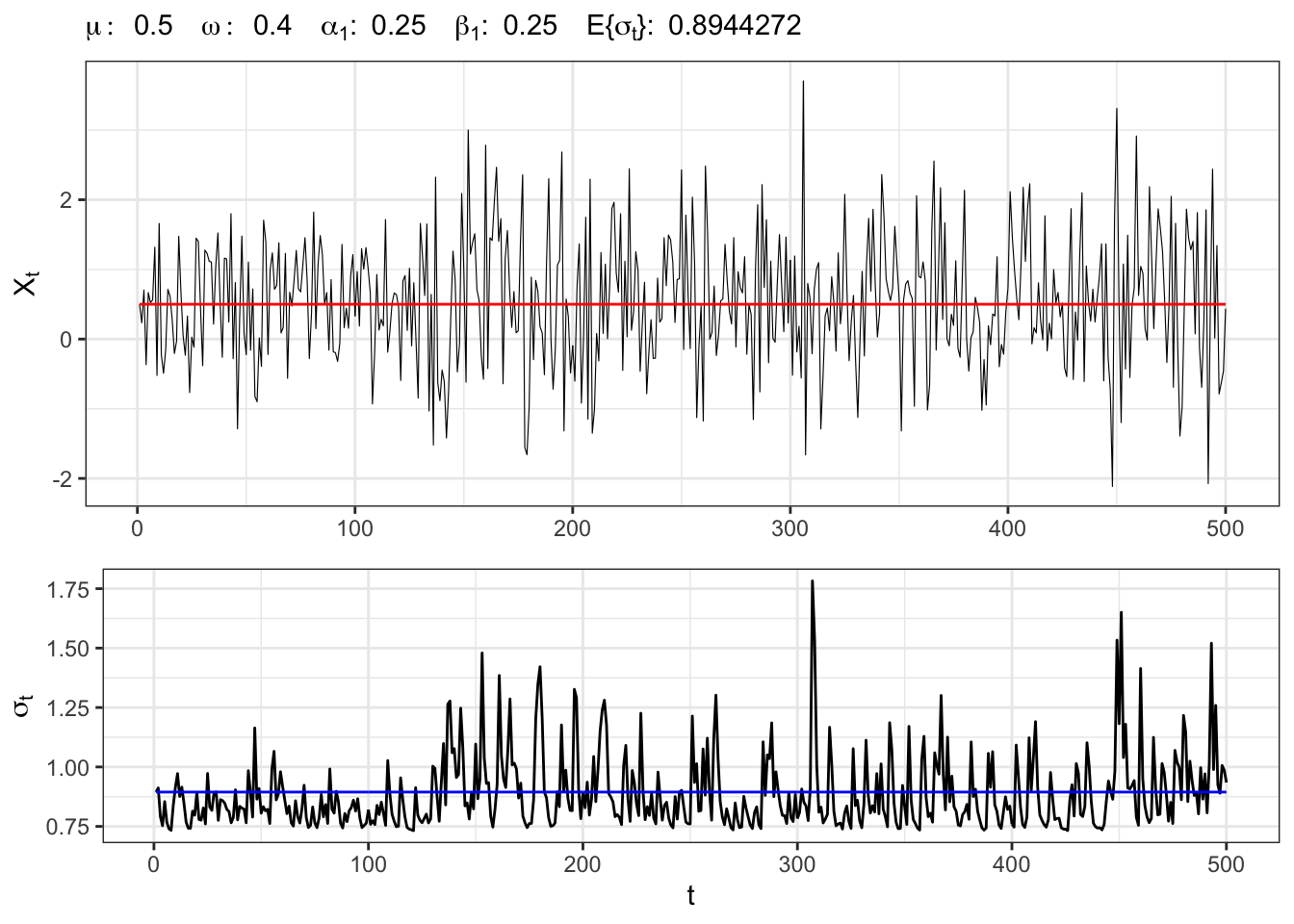
2.1 Example: GARCH(1,1) process
Let’s simulate an GARCH(1,1) process with normal residuals, namely
GARCH(1,1) simulation
# Random seed
set.seed(3)
# ==================== Setups =====================
# number of simulations
t_bar <- 500
# parameters
par <- c(mu=0.5, omega=0.4, alpha1=0.25, beta1=0.25)
# long-term std deviation of residuals
sigma_eps <- sqrt(par[2]/(1 - par[3] - par[4]))
# ================== Simulation ===================
# Initial points
Xt <- rep(par[1], 1)
sigma <- rep(sigma_eps, 1)
# Simulated residuals
eps <- rnorm(t_bar, 0, 1)
for(t in 2:t_bar){
# GARCH(1,1) variance
sigma[t] <- par[3]*eps[t-1]^2 + par[4]*sigma[t-1]^2
# GARCH std. deviation
sigma[t] <- sqrt(par[2] + sigma[t])
# Simulated residuals
eps[t] <- sigma[t]*eps[t]
# Simulated time series
Xt[t] <- par[1] + eps[t]
}
# ===================== Plot ======================
# GARCH(1,1) simulation
plot_garch <- ggplot()+
geom_line(aes(1:t_bar, Xt), size = 0.2)+
geom_line(aes(1:t_bar, par[1]), color = "red")+
theme_bw()+
labs(x = NULL, y = TeX("$X_t$"),
subtitle = TeX(paste0("$\\mu:\\;", par[1],
"\\;\\; \\omega:\\;", par[2],
"\\;\\; \\alpha_{1}:\\;", par[3],
"\\;\\; \\beta_{1}:\\;", par[4],
"\\;\\; E\\{\\sigma_t\\}:\\;", sigma_eps,
"$")))
# GARCH(1,1) std. deviation
plot_sigma <- ggplot()+
geom_line(aes(1:t_bar, sigma), alpha = 1)+
geom_line(aes(1:t_bar, sigma_eps), alpha = 1, color = "blue")+
labs(x = "t", y = TeX("$\\sigma_{t}$"))+
theme_bw()
gridExtra::grid.arrange(plot_garch, plot_sigma, heights = c(0.6, 0.4))2.2 Example: GARCH(2,3) process
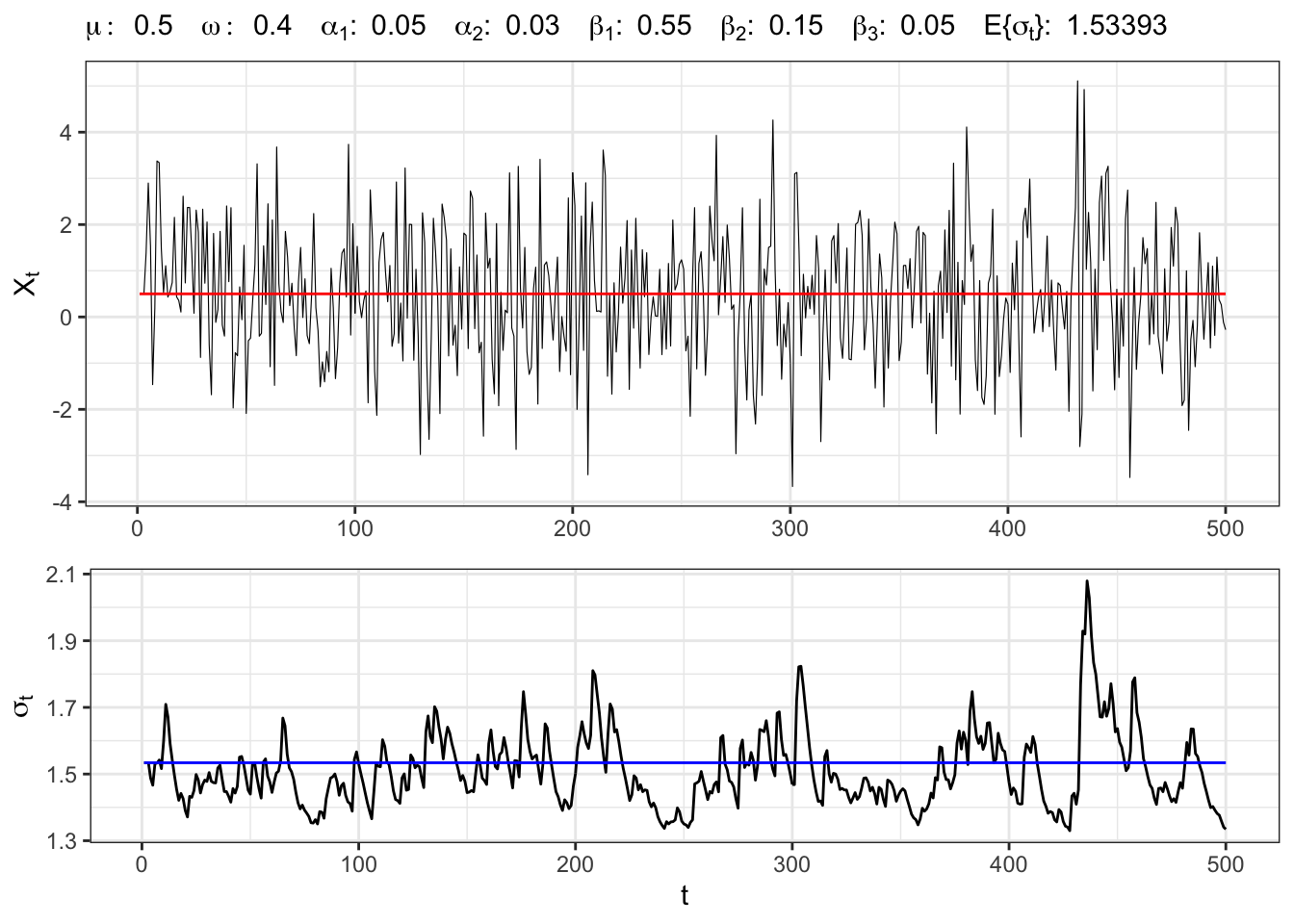
Let’s simulate an GARCH(2,3) process with normal residuals, namely
GARCH(2,3) simulation
# Random seed
set.seed(4)
# ==================== Setups =====================
# Number of simulations
t_bar <- 500
# Parameters
par <- c(mu=0.5, omega=0.4,
alpha1=0.05, alpha1=0.03,
beta1=0.55, beta2=0.15, beta3=0.05)
# Long-term std deviation of residuals
sigma_eps <- sqrt(par[2]/(1 - sum(par[3:7])))
# ================== Simulation ===================
# Initial points
Xt <- rep(par[1], 4)
sigma <- rep(sigma_eps, 4)
# Simulated residuals
eps <- rnorm(t_bar, 0, 1)
for(t in 4:t_bar){
# ARCH(2) variance
sigma2_arch <- par[3]*eps[t-1]^2 + par[4]*eps[t-2]^2
# GARCH(3) variance
sigma2_garch <- par[5]*sigma[t-1]^2 + par[6]*sigma[t-2]^2 + par[7]*sigma[t-3]^2
# GARCH(2,3) std. deviation
sigma[t] <- sqrt(par[2] + sigma2_arch + sigma2_garch)
# Simulated residuals
eps[t] <- sigma[t]*eps[t]
# Simulated time series
Xt[t] <- par[1] + eps[t]
}
# ===================== Plot ======================
# GARCH(2,3) simulation
plot_garch <- ggplot()+
geom_line(aes(1:t_bar, Xt), size = 0.2)+
geom_line(aes(1:t_bar, par[1]), color = "red")+
theme_bw()+
labs(x = NULL, y = TeX("$X_t$"),
subtitle = TeX(paste0("$\\mu:\\;", par[1],
"\\;\\; \\omega:\\;", par[2],
"\\;\\; \\alpha_{1}:\\;", par[3],
"\\;\\; \\alpha_{2}:\\;", par[4],
"\\;\\; \\beta_{1}:\\;", par[5],
"\\;\\; \\beta_{2}:\\;", par[6],
"\\;\\; \\beta_{3}:\\;", par[7],
"\\;\\; E\\{\\sigma_t\\}:\\;", sigma_eps,
"$")))
# GARCH(2,3) std. deviation
plot_sigma <- ggplot()+
geom_line(aes(1:t_bar, sigma), alpha = 1)+
geom_line(aes(1:t_bar, sigma_eps), color = "blue")+
labs(x = "t", y = TeX("$\\sigma_{t}$"))+
theme_bw()
gridExtra::grid.arrange(plot_garch, plot_sigma, heights = c(0.6, 0.4))2.3 Example: GARCH(3,2) process
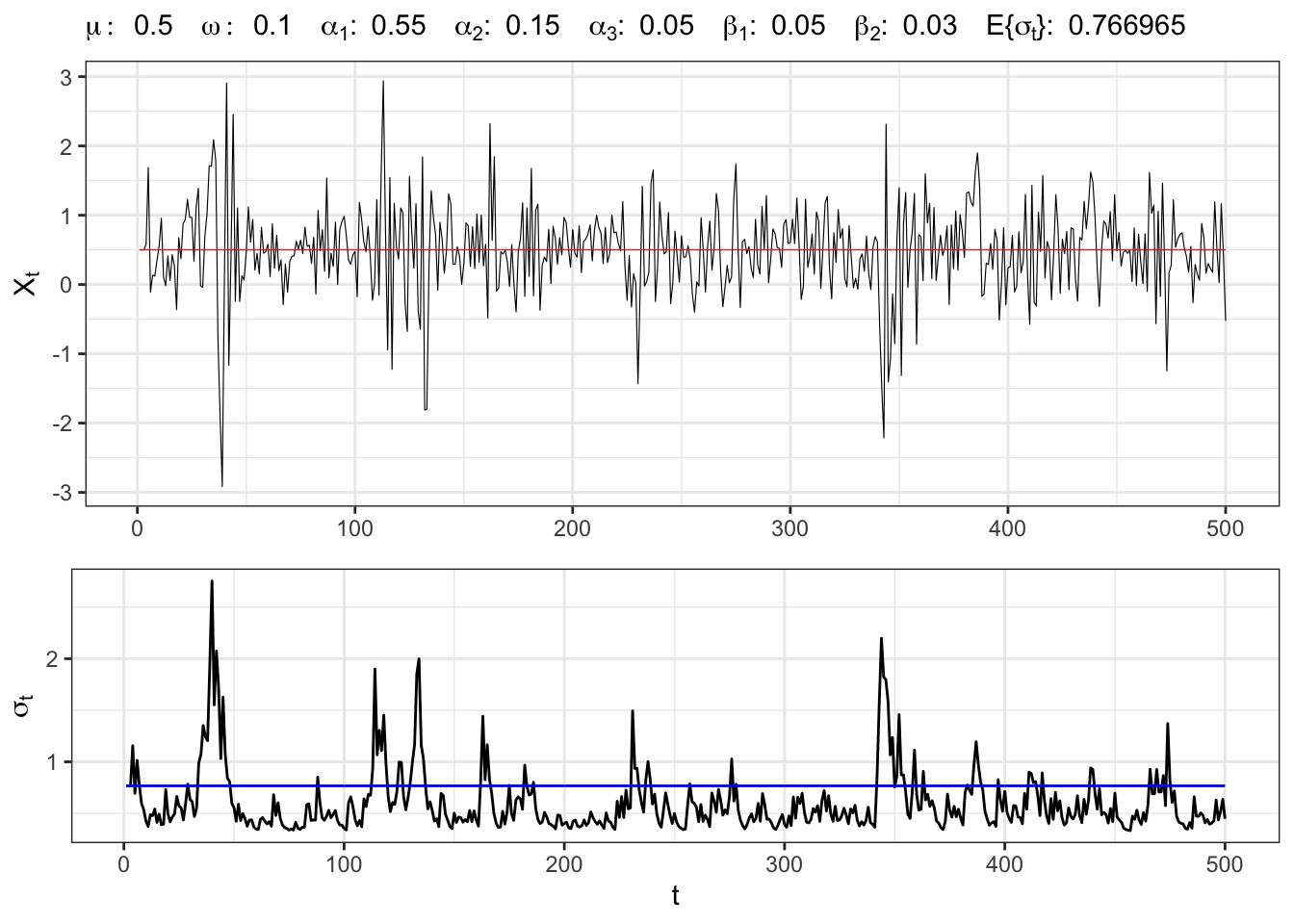
Let’s simulate an GARCH(3,2) process with normal residuals, namely
GARCH(3,2) simulation
# Random seed
set.seed(5)
# ==================== Setups =====================
# Number of simulations
t_bar <- 500
# Parameters
par <- c(mu = 0.5, omega = 0.1, alpha1 = 0.55,
alpha2 = 0.15, alpha3 = 0.05,
beta1 = 0.05, beta2 = 0.03)
# Long-term std deviation of residuals
sigma_eps <- sqrt(par[2]/(1 - sum(par[3:7])))
# ================== Simulation ===================
# Initial points
Xt <- rep(par[1], 4)
sigma <- rep(sigma_eps, 4)
# Simulated residuals
eps <- rnorm(t_bar, 0, 1)
for(t in 4:t_bar){
# ARCH(3) variance
sigma2_arch <- par[3]*eps[t-1]^2 + par[4]*eps[t-2]^2 + par[5]*eps[t-3]^2
# GARCH(2) variance
sigma2_garch <- par[6]*sigma[t-3]^2 + par[7]*sigma[t-2]^2
# GARCH(3,2) std. deviation
sigma[t] <- sqrt(par[2] + sigma2_arch + sigma2_garch)
# Simulated residuals
eps[t] <- sigma[t]*eps[t]
# Simulated time series
Xt[t] <- par[1] + eps[t]
}
# ===================== Plot ======================
# GARCH(3,2) simulation
plot_garch <- ggplot()+
geom_line(aes(1:t_bar, Xt), size = 0.2)+
geom_line(aes(1:t_bar, par[1]), color = "red", size = 0.2)+
theme_bw()+
labs(x = NULL, y = TeX("$X_t$"),
subtitle = TeX(paste0("$\\mu:\\;", par[1],
"\\;\\; \\omega:\\;", par[2],
"\\;\\; \\alpha_{1}:\\;", par[3],
"\\;\\; \\alpha_{2}:\\;", par[4],
"\\;\\; \\alpha_{3}:\\;", par[5],
"\\;\\; \\beta_{1}:\\;", par[6],
"\\;\\; \\beta_{2}:\\;", par[7],
"\\;\\; E\\{\\sigma_t\\}:\\;", sigma_eps,
"$")))
# GARCH(3,2) std. deviation
plot_sigma <- ggplot()+
geom_line(aes(1:t_bar, sigma), alpha = 1)+
geom_line(aes(1:t_bar, sigma_eps), color = "blue")+
labs(x = "t", y = TeX("$\\sigma_{t}$"))+
theme_bw()
gridExtra::grid.arrange(plot_garch, plot_sigma, heights = c(0.6, 0.4))3 IGARCH
Integrated Generalized Autoregressive Conditional heteroskedasticity (IGARCH(p,q)) is a restricted version of the GARCH model, where the persistent parameters sum up to one, and imports a unit root in the GARCH process. The condition for this is
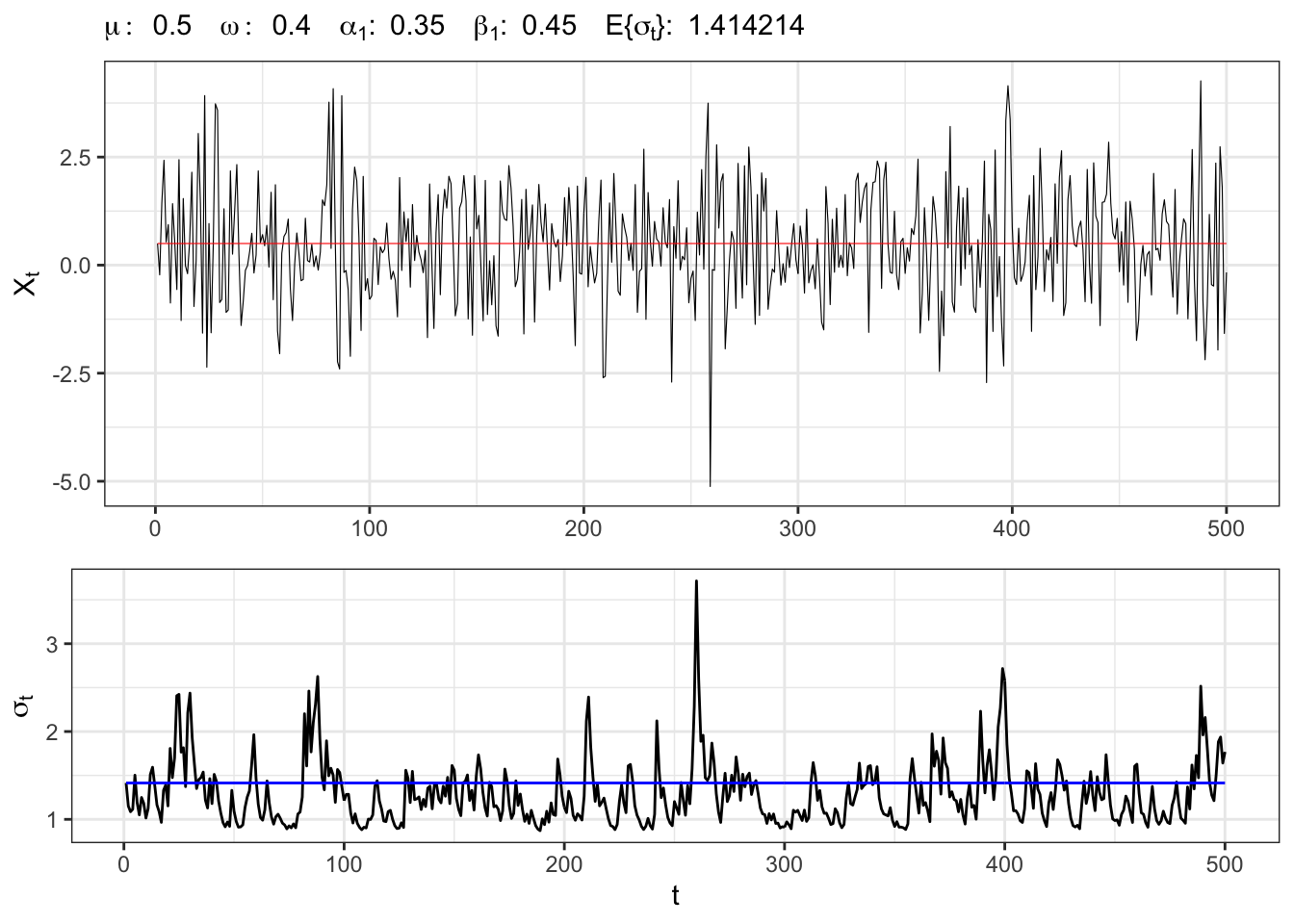
3.1 Example: IGARCH(1,1)
IGARCH(1,1) simulation
# Random seed
set.seed(6)
# ==================== Setups =====================
# number of simulations
t_bar <- 500
# parameters
par <- c(mu=0.5, omega=0.4, alpha1=0.35, beta1=0.45)
# long-term std deviation of residuals
sigma_eps <- sqrt(par[2]/(1 - par[3] - par[4]))
# ================== Simulation ===================
# Initial points
Xt <- rep(par[1], 1)
sigma <- rep(sigma_eps, 1)
# Simulated residuals
eps <- rnorm(t_bar, 0, 1)
for(t in 2:t_bar){
# iGARCH(1,1) variance
sigma[t] <- par[3]*eps[t-1]^2 + par[4]*sigma[t-1]^2
# iGARCH std. deviation
sigma[t] <- sqrt(par[2] + sigma[t])
# Simulated residuals
eps[t] <- sigma[t]*eps[t]
# Simulated time series
Xt[t] <- par[1] + eps[t]
}
# ===================== Plot ======================
# iGARCH(1,1) simulation
plot_garch <- ggplot()+
geom_line(aes(1:t_bar, Xt), size = 0.2)+
geom_line(aes(1:t_bar, par[1]), color = "red", size = 0.2)+
theme_bw()+
labs(x = NULL, y = TeX("$X_t$"),
subtitle = TeX(paste0("$\\mu:\\;", par[1],
"\\;\\; \\omega:\\;", par[2],
"\\;\\; \\alpha_{1}:\\;", par[3],
"\\;\\; \\beta_{1}:\\;", par[4],
"\\;\\; E\\{\\sigma_t\\}:\\;", sigma_eps,
"$")))
# iGARCH(1,1) std. deviation
plot_sigma <- ggplot()+
geom_line(aes(1:t_bar, sigma), alpha = 1)+
geom_line(aes(1:t_bar, sigma_eps), alpha = 1, color = "blue")+
labs(x = "t", y = TeX("$\\sigma_{t}$"))+
theme_bw()
gridExtra::grid.arrange(plot_garch, plot_sigma, heights = c(0.6, 0.4))4 GARCH-M
The GARCH in-mean (GARCH-M) model adds a stochastic term into the mean equation, i.e.
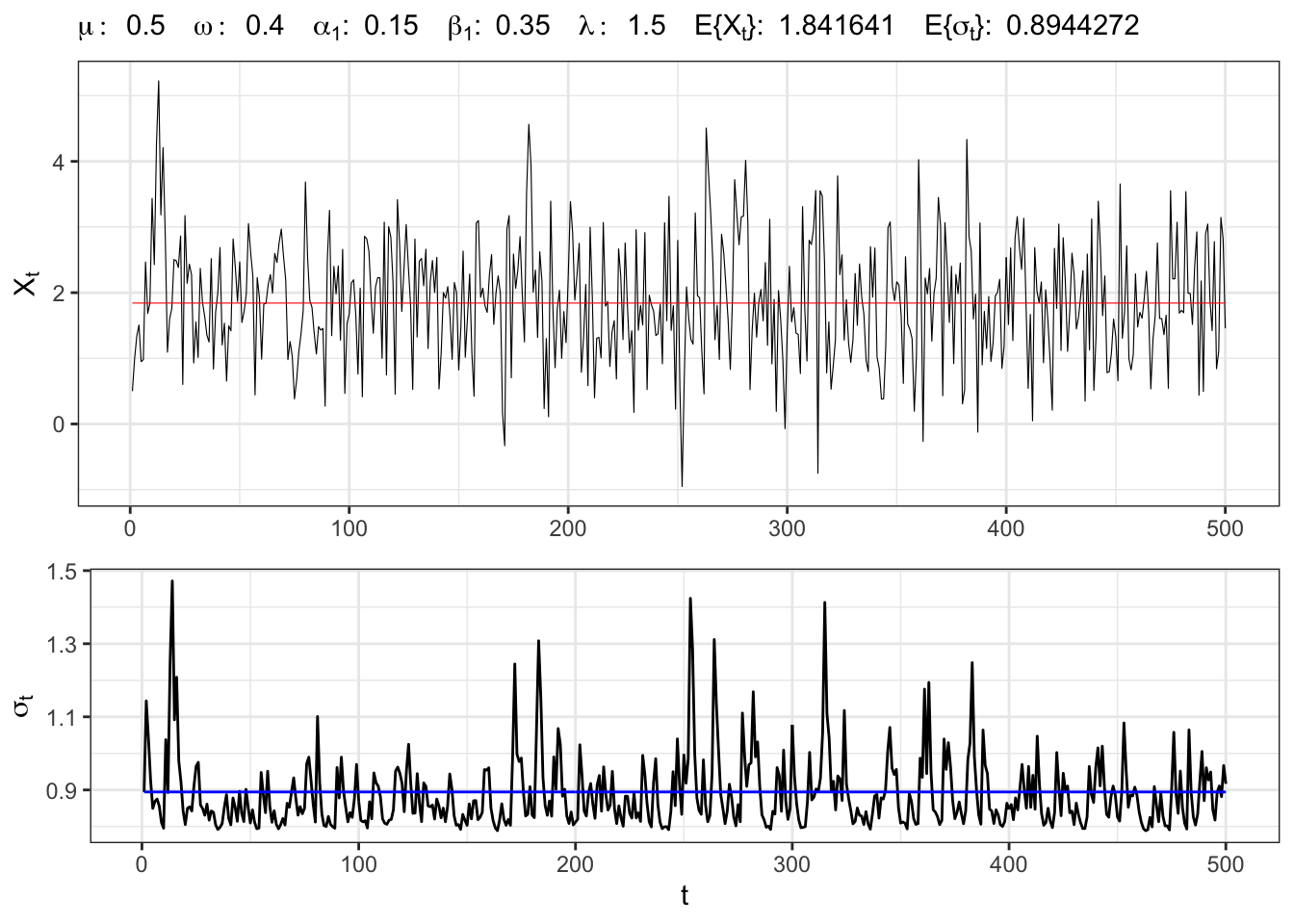
4.1 Example: GARCH-M(1,1)
GARCH-M(1,1) simulation
set.seed(7)
# ==================== Setups =====================
# number of simulations
t_bar <- 500
# parameters
par <- c(mu=0.5, omega=0.4,
alpha1=0.15, beta1=0.35, lambda = 1.5)
# long-term std deviation of residuals
sigma_eps <- sqrt(par[2]/(1 - par[3] - par[4]))
# ================== Simulation ===================
# Initial points
Xt <- rep(par[1], 1)
sigma <- rep(sigma_eps, 1)
# Simulated residuals
eps <- rnorm(t_bar, 0, 1)
eps[1:2] <- eps[1:2]*sigma_eps
for(t in 2:t_bar){
# GARCH(1,1) variance
sigma[t] <- par[3]*eps[t-1]^2 + par[4]*sigma[t-1]^2
# GARCH std. deviation
sigma[t] <- sqrt(par[2] + sigma[t])
# Simulated residuals
eps[t] <- sigma[t]*eps[t]
# Simulated time series
Xt[t] <- par[1] + sigma[t]*par[5] + eps[t]
}
# ===================== Plot ======================
# GARCH-M(1,1) simulation
plot_garch <- ggplot()+
geom_line(aes(1:t_bar, Xt), size = 0.2)+
geom_line(aes(1:t_bar, par[1] + sigma_eps*par[5]), color = "red", size = 0.2)+
theme_bw()+
labs(x = NULL, y = TeX("$X_t$"),
subtitle = TeX(paste0("$\\mu:\\;", par[1],
"\\;\\; \\omega:\\;", par[2],
"\\;\\; \\alpha_{1}:\\;", par[3],
"\\;\\; \\beta_{1}:\\;", par[4],
"\\;\\; \\lambda:\\;", par[5],
"\\;\\; E\\{\\X_t\\}:\\;", par[1] + sigma_eps*par[5],
"\\;\\; E\\{\\sigma_t\\}:\\;", sigma_eps,
"$")))
# GARCH-M(1,1) std. deviation
plot_sigma <- ggplot()+
geom_line(aes(1:t_bar, sigma), alpha = 1)+
geom_line(aes(1:t_bar, sigma_eps), alpha = 1, color = "blue")+
labs(x = "t", y = TeX("$\\sigma_{t}$"))+
theme_bw()
gridExtra::grid.arrange(plot_garch, plot_sigma, heights = c(0.6, 0.4))Citation
@online{sartini2024,
author = {Sartini, Beniamino},
title = {ARCH-GARCH Process},
date = {2024-05-01},
url = {https://greenfin.it/statistics/arch-garch-process.html},
langid = {en}
}